- Researchers using DNA barcoding technology found that over 70% of shark fins and ray gill plates, collected from sellers in multiple countries, came from threatened species.
- They determined the species of 129 dried commercial fin and gill samples, which are otherwise difficult to identify, by comparing genetic material in the samples with samples in two reference databases.
- The study’s findings support the use of DNA barcoding as a tool to help enforcement agencies determine whether processed specimens derive from legal or illegal species.
- Their identification of at least 20 shark and 5 ray species, 12 of which are under trade restrictions, suggests that current global shark and ray fishing is unsustainable and merits a strong conservation response.
Markets around the world sell products made from the processed parts of wild animals and plants, which are sometimes illegal and mislabeled.
A research team using DNA barcoding technology to identify shark fins and manta gills sold commercially found that a majority of samples they tested, from sellers in multiple countries, were from threatened species.
Their findings show that DNA barcoding holds potential to help enforcement agencies identify illegal products that in hand cannot be distinguished from legal imports. The success of the method should also help researchers better determine population status of species of conservation concern.

Populations of many shark and ray species have declined dramatically, shrinking by up to 85% in less than 10 years. Trade data from Asian markets suggest that 26–73 million sharks are caught each year for the fin trade alone.
The high price of shark fin soup and wildlife products for Chinese medicine and huge demand for these among Chinese customers encourages fishers to target the fins as a luxury commodity. Fishing fleets from Mexico to Mozambique to Indonesia kill large numbers of sharks, slice off their fins, and toss the rest of each now-doomed animal back into the sea to save space on their vessels for more fins.

A new trend in Chinese medicine is the consumption of the gills of various ray species to boost the immune system and cure a range of ailments. However, like shark fins, the gills of manta and mobula rays are made of cartilage and have no known medicinal value.
The killing occurs across the broad geographic ranges of these animals and now threatens 25% of shark and ray species, collectively called elasmobranchs, a branch of cartilaginous fishes.
Determining the sustainability of elasmobranch fisheries worldwide has eluded scientists and resource managers. The processed body parts—namely dried shark fins and ray gill plates—of different species look very similar, and common names used in their trade rarely correspond to their taxonomic names.
Some shark and ray fishing is legal, say the researchers, but the number of animals killed by illegal, unreported and unregulated (IUU) fisheries or as undeclared by-catch “likely far outweighs those from legal fisheries.”
A better understanding of the population status of these species will require major improvements to species catch data records, which, in turn, requires better species identification.
New tech may compensate for a lack of traditional species indicators
The researchers, from University of Guelph and Nova Southeastern University, obtained 129 samples from various sources. Their samples included 71 dried shark fins from retailers in Vancouver in Canada, 54 ray gill plates from retailers in Hong Kong and mainland China, and four additional gill plates from fisheries in Sri Lanka.
 |
 |
| Two of the market samples analyzed in the study and added to the BOLD data base. On left, the dried fin of a blue shark from Vancouver, Canada. On the right, the dried fin of a bigeye thresher shark, also from Vancouver. Image credits: Steinke et al, 2017. | |
To determine the species of these products, they analyzed the DNA barcode sequences derived from the fin samples on the Barcode of Life Data System’s (BOLD) cloud-based data storage and analysis platform. The BOLD data base is freely available for any researcher interested in DNA barcoding to acquire, store, analyze, and publish DNA barcode records. It houses genetic data, combined with information on morphology and distribution, for nearly 200,000 animal species.
DNA barcoding uses a genetic fragment of roughly 650 base pairs—the “barcode”—that is taken from a standard position in an animal’s genome. Barcodes have been effective in identifying animal DNA as belonging to a particular species; they thus allow scientists to identify organisms and specimens using genetic material.

The researchers used the BOLD platform’s “species”-level identification function to compare the barcode of each sample against the barcodes from voucher specimens that had been identified by experts. The platform successfully matched each sample with a species in the BOLD database (similarity of at least 99%). The researchers obtained the same level of species-level sequence similarity for the gill samples using another barcode database service (BLAST) at the National Center for Biotechnology Information (NCBI).
Air-dried fish samples typically degrade DNA, but even the shorter intact sequences (93-184 base pairs) tested in this study contained enough variation for successful identification of the shark fins to species.
“DNA barcoding is an ideal tool when identifying dried samples or samples that have been processed,” said the study’s lead author Dirk Steinke of the University of Guelph in a media release. “It provides enforcement agencies with a method for detecting whether the fins and gills that are being sold are legal or illegal imported species.”

The barcode sequences from the 41 ray gill plate samples were sufficient to distinguish them by genus but not to unambiguously identify the samples to species level. The study explains that uncertainty remains about the separation of the Mobula (devil ray) genus into two distinct species. In addition, the two Manta species are thought to have separated recently and may continue to hybridize, keeping their DNA more similar. Nevertheless, both Manta species are now protected in international waters and their trade restricted by the Convention for the International Trade in Endangered Species (CITES).

The researchers contributed their information on the species and the samples, including where and when each was collected, in the public dataset DS-SHMA (dx.doi.org/10.5883/DS-SHMA) on the BOLD data storage platform. To continue expanding the reference database, they deposited samples in GenBank, the National Institute of Health’s free, public, online repository for genetic/genomic data.
Species of conservation concern
The study identified parts from at least 20 shark and 5 ray species, 12 of which are now under trade restrictions. The researchers detected both fin and gill plate samples from the whale shark, an endangered species. Whale sharks are listed on Appendix II of CITES since 2003, requiring close governmental control of its trade, suggesting the samples were likely illegal.
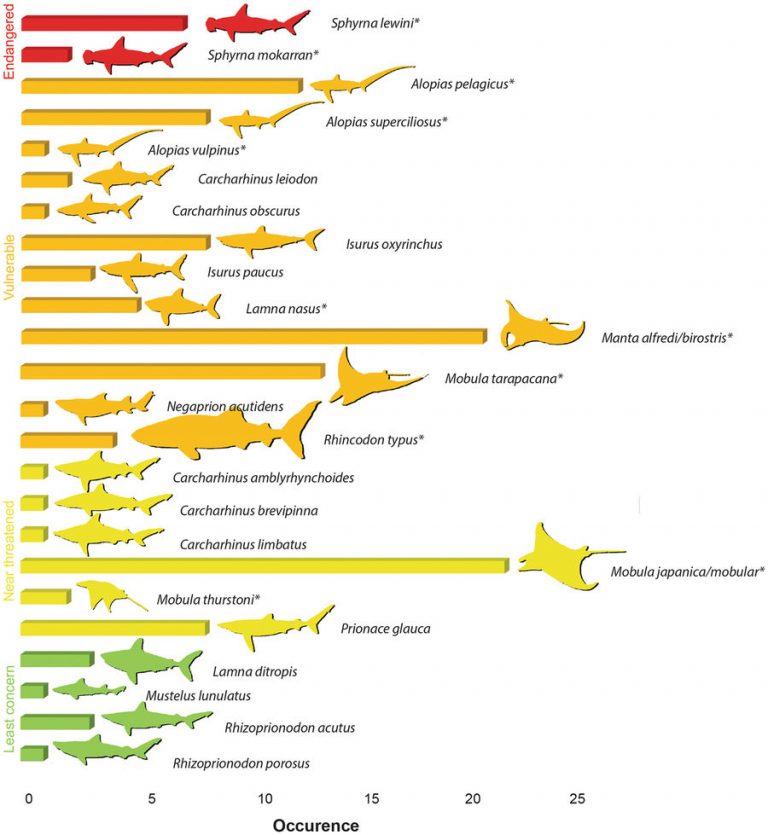
Moreover, the slow growth, onset of breeding later in life, and slow reproduction of many elasmobranch species reduce their ability to recover population numbers once they are depleted. This study’s finding, therefore, that 71% of the fin and gill plate samples are from these species of high conservation concern suggests that the current fishing practices globally are highly unsustainable.
In their paper, the researchers urge “an extensive conservation management response” that includes large-scale long-term surveillance of markets, as well as of fisheries and marine protected areas. As DNA analysis becomes both more portable and able to identify species from the small, dried specimens typically found in markets and transportation ports, such surveillance of wildlife products along their supply chains will become increasingly achievable.
Reference
Steinke, D., Bernard, A. M., Horn, R. L., Hilton, P., Hanner, R., & Shivji, M. S. (2017). DNA analysis of traded shark fins and mobulid gill plates reveals a high proportion of species of conservation concern. Scientific reports, 7(1), 9505.







