- Environmental DNA, known as eDNA, is genetic material sloughed off by animals or plants and found in soil, air, or water, and allows scientists to collect and analyze genetic material without having to retrieve it from a species directly.
- Researchers in Hawaii found that the amount of eDNA in water samples is related to coral abundance and thus can be used to conduct accurate surveys of local coral populations using less time and money than sending SCUBA divers down to do the surveys.
- Coral reefs have the highest biodiversity of any ecosystem globally and are one of the most threatened, thanks to climate change and direct human impact. eDNA could help researchers evaluate coral abundance and health more quickly, easily and cost-effectively.
Faster and cheaper
Surveying and studying coral takes a lot of work. It’s usually done manually, which requires wet suits and air tanks and SCUBA gear and people. But it’s important to do, as coral reefs have the most biodiversity of any ecosystem on Earth yet are degrading rapidly due to rising sea temperatures.
Collecting water samples is much simpler and cheaper. And now a recent study shows that the eDNA found in these samples can give scientists the same accurate survey information without sending any divers down into the deep blue sea.

eDNA, short for environmental DNA, is the DNA found in the bits of biological residue in soil, air, and water. In the past, collecting DNA from a certain plant or animal required scientists to physically come into contact with the species and take a sample directly. With eDNA however, scientists are able to collect DNA from the feces, pollen, skin, scales, or mucus found in soil or water samples.
According to new research from the University of Hawai`i at Mānoa Department of Biology, eDNA can be used to accurately survey the amount of living coral in reefs. This study, led by Patrick Nichols, a marine biology graduate student, and associate professor Peter Marko, found a high correlation between the amount of living coral counted using visual surveys and the concentration of coral eDNA found in samples of seawater. Although there is a growing body of research being done with eDNA, few studies have shown reliable links between organism abundance and presence of eDNA. This study shows that eDNA can be a quick and affordable way to measure live coral presence in a reef.

The technique used to extract and evaluate eDNA from the seawater, called metabarcoding, analyzes all the DNA in a water sample with DNA primers to amplify specific sections of DNA from target organisms in order to identify which are present in the sample. Some of the target primer sequences in the current study were short, amplifying only about 80-120 base pairs of DNA, while the longest sequences had around 400. These primers, designed to amplify DNA of all coral genera found in the Hawaiian Islands, were able to not only quantify coral cover but also detect differences in coral community structure (i.e., the relative abundances of major coral genera) at different sites from the eDNA in the water samples.
According to Nichols, coral is a good candidate for this kind of research because there are plenty of coral sequences that exist on public online databases, where they can be matched with newly collected eDNA sequences. “Most coral species, at least the genus, will have some sort of reference,” Nichols told Mongabay. “Even with an extremely small fragment of DNA, we can still get an idea of what the organism is.”
The research involved collecting samples of seawater from a range of sites on the island of Oahu with varying levels of coral degradation. Some of the sites were marine reserves, some were touristy beaches, and one had less than 1 percent coral cover overall. Once researchers collected the water, they had to quickly extract the DNA and stabilize it, because otherwise it would degrade quickly and become unusable. Nichols and his team then put the DNA through a number of filters to sift out bacteria and other single-celled organisms that make up much of the DNA in the water, as they were only interested in larger, multi-celled organisms. They then compared the filtered DNA with online databases to match it up with known sequences.

Healthy coral shed a lot of eDNA, while degraded reefs left very little in the water, giving information about which corals in which areas were experiencing more damage. Nichols and Marko found their eDNA results to be highly correlated with those found during visual surveys done by SCUBA divers in the same areas.
The challenges of eDNA
There are still many questions when it comes to data resulting from the use of environmental DNA. One challenge for this type of eDNA study is that water is constantly flowing, which makes it difficult to know where the DNA in a particular sample actually came from.
“There is a need to be extremely aware of where the water is going and where the water is coming from,” said Nichols. “Most of our sites are open, coastal areas, so there’s not too much current.” While the effect of water flows has yet to be quantified extensively, the general consensus according to Nichols is that because DNA degrades so rapidly in a marine environment, the site where the sample is taken most likely contains eDNA from the same general area.
Another challenge is that the biomass of each group of organisms at a site should theoretically be represented in the proportion of their eDNA found in a water sample from that site. This, however, presupposes that all these organisms are shedding their DNA at roughly the same rate, which is not necessarily the case. Different species will shed mucus at different rates, certain fish poop more than others, and the abundance of reproductive material in the water will change seasonally. “If you’re looking at a whole community,” Nichols said, “you need those rates to all be similar to make assumptions about abundance.”
Certain organisms and environments are better suited for using eDNA to conduct population surveys than others. The surface area of coral species, for example, is a good proxy for their actual biomass, and it is reasonable to assume that all coral species shed at similar rates, making coral a good study subject for eDNA surveying. And because coral is such a vital part of reef ecosystems, coral cover is an important marker of overall reef health, making it an important organism to study.

Due to rising water temperatures from global warming, along with direct human impacts like pollution, coral cover worldwide is degrading rapidly. Over the last five years, there have been mass coral bleaching events due to the rise in ocean temperature. According to UNESCO, the coral in all 29 World Heritage sites that contain reefs could cease to exist by the end of this century if greenhouse gas emissions continue at their current rate. Reefs support more than 4,000 fish species globally and provide food, coastline protection, and incomes for millions of people worldwide, so it is important to be able to monitor and survey reef degradation, which eDNA analysis can make easier and more affordable to do.
Looking ahead
The study’s findings suggest that other sites could monitor coral cover and biomass through rapid surveys using relatively small volumes of seawater and that the new method could assist with existing monitoring efforts, especially where visual surveys by divers are logistically challenging.
Nichols said he plans to continue monitoring coral to build a long-term large-scale data set, including areas before and after bleaching events. He added that he would also like to focus on deep-water reefs, those around 60–200 meters (197–656 feet) down that are shallow enough not to need submersibles but too deep for recreational diving. That area, he says, is insufficiently studied, so he would like to use eDNA to fill in that knowledge gap.
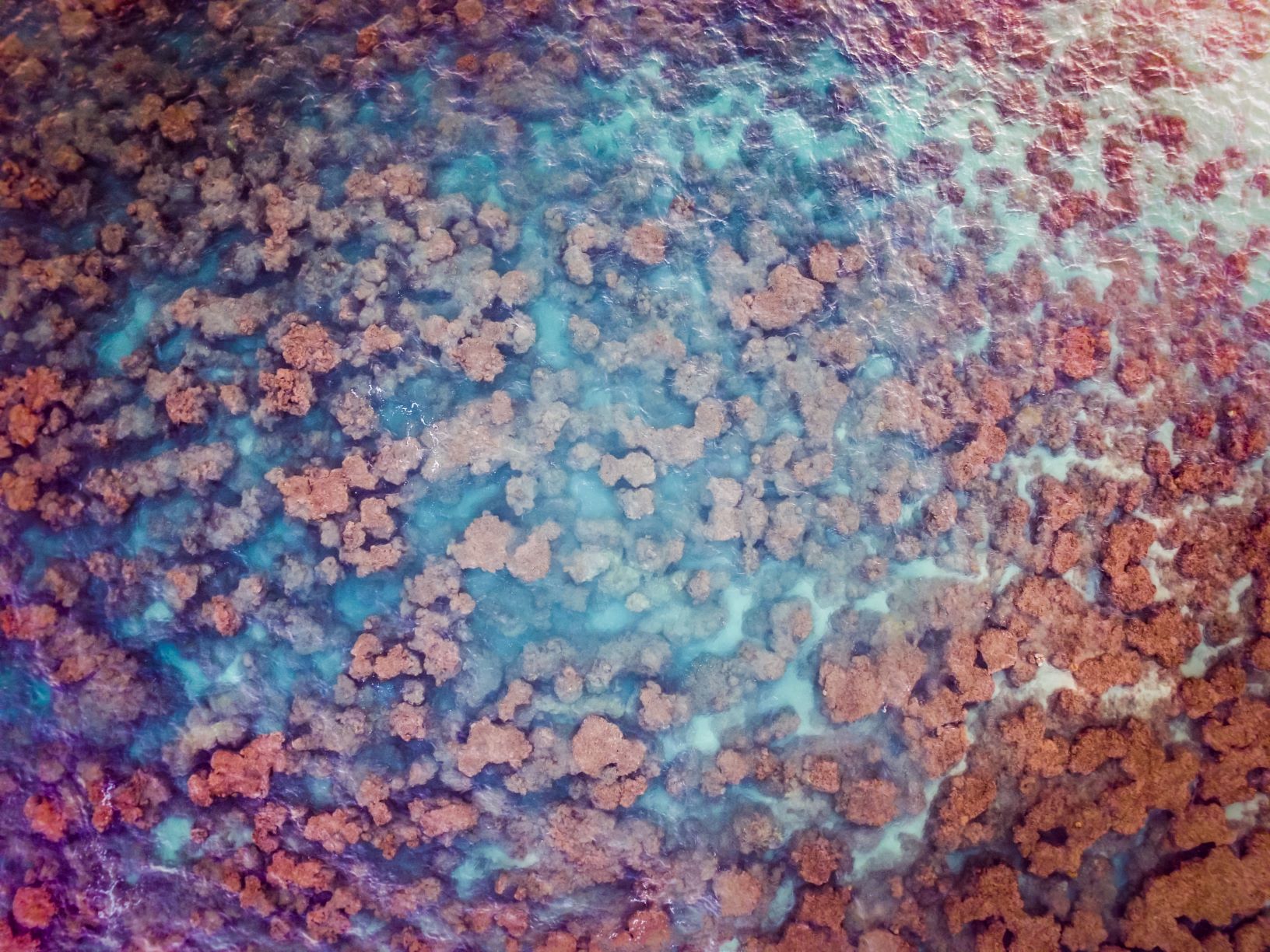
eDNA already promises coral surveys that are cheaper and faster and potentially in areas otherwise hard to access, but Nichols says there is still a lot to be done to make eDNA research better. “Right now, it seems pretty haphazard how people are choosing their primers,” Nichols said. He would like to see some standardization of the research methods. In addition, obstacles such as quantifying water flow and determining how much DNA particular species shed will need to be addressed in the coming years.
“The big take home is that it’s a lot cheaper than the methods currently being used,” Nichols said. “It’s a lot less money than hiring a team of divers.”
Citation
Nichols, P. K. & Marko, P. B. (2019). Rapid assessment of coral cover from environmental DNA in Hawai’i. Environmental DNA, 1-14.
Banner image of a diversity of corals by Sue Palminteri/Mongabay.
FEEDBACK: Use this form to send a message to the editor of this post. If you want to post a public comment, you can do that at the bottom of the page.
